How to extract digitizer peaks with the SCS Toolbox¶
Workflow during a beamtime¶
Record first data with signal on digitizer.
Find peak integration parameters using
check_peak_params().Update the new parameters in DAMNIT, so that for each new run, automatic processing of each run is performed and saved in
usr/processed_runsfolder. As long as the right bunch pattern is selected for peak extraction, there is no need to care about the number of pulses / period, as they will be adjusted to match the bunch pattern of each run.For analysis, load digitizer data using
load_processed_peaks()orload_all_processed_peaks(). This is much faster than loading the raw traces and re-performing peak integration.Checking the integration parameters used for the processed data can be done via
check_processed_peaks_params().
Peak-integration parameters¶
[1]:
import toolbox_scs as tb
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['figure.constrained_layout.use'] = True
Cupy is not installed in this environment, no access to the GPU
Extracting peaks from a raw trace is done using the get_digitizer_peaks() function of the SCS Toolbox, by integration over the area of the peak and subtraction of a baseline. For one peak, the parameters pulseStart and pulseStop (sample numbers in the raw trace) define the integration region and baseStart and baseStop define the baseline region. In most cases, the pulse pattern is regular and there are npulses separated by a period. The peak extraction is repeated for
each peak, with pulseStart, pulseStop, baseStart and baseStop shifted by the period.
An example of integration parameters:
[2]:
params = {'pulseStart': 100,
'pulseStop': 120,
'baseStart': 80,
'baseStop': 99,
'period': 96,
'npulses': 25}
If the pattern is not regular, a list of starting positions can be provided to pulseStart, while pulseStop, baseStart, baseStop remain integers and relate to the first peak only. In such case, period does not have a meaning and npulses is equal to len(pulseStart):
[3]:
params = {'pulseStart': [100, 200, 500, 600, 900, 1000, 2000, 10000, 15500],
'period': 0,
'pulseStop': 110,
'baseStop': 99,
'baseStart': 90,
'npulses': 9}
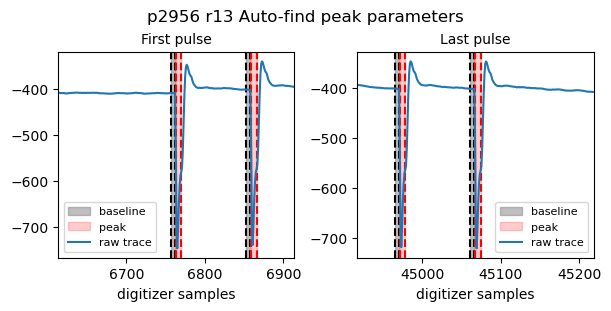
Let’s assume that we are interested in the APD signal on diode 8 looking at the FEL, which corresponds to Ch9 of Fast ADC 2 (mnemonic FastADC2_9raw). We can check how the peak-finding algorithm performs by using tb.check_peak_parameters() and inspecting the found regions of integration. This shows a plot with the first and last pulses identified by the peak-finding algorithm and displays the region of integration and the region for baseline subtraction. The function returns a dictionnary
good_params that has all parameters necessary to perform the trapezoidal integration over the digitizer trace.
[4]:
proposal, runNB = 2956, 13
good_params = tb.check_peak_params(proposal, runNB, 'FastADC2_9raw', bunchPattern='sase3')
good_params
Bunch pattern sase3: 400 pulses, 96 samples between two pulses
Auto-find peak parameters: 400 pulses, 96 samples between two pulses
[4]:
{'pulseStart': 6763,
'period': 96,
'pulseStop': 6770,
'baseStop': 6762,
'baseStart': 6757,
'npulses': 400}
Extracting peaks¶
The integration parameters are either user-provided or automatically computed by a peak-finding algorithm in get_digitizer_peaks() and check_peak_params(). The bunch pattern, when provided, is used to determine the parameters or to check consistency with user-provided parameters, and to align the pulse ID. The minimum required inputs to extract peaks are:
the
bunchPatternsource (‘sase3’ if the device is looking at the FEL or ‘scs_ppl’ if the device is looking at the PP laser), leavingintegParams=Noneto let the peak-finding algorithm operate.or
integParamsdict includingpulseStart,pulseStop,baseStart,baseStop,periodandnpulseskeys.
In most cases, automatic peak finding provides good integration parameters. If it fails, or if we want to define fixed parameters to consistently analyze a series of runs, it is necessary to provide the parameters via integParams.
If both the bunch pattern and the integration parameters are provided, the period and npulses of the user-provided parameters (integParams) will be overriden (with a warning in case of mismatch), except if pulseStart is a list (e.g. case of irregular patterns).
Once the parameters are found, we can extract the peaks using get_digitizer_peaks(), with bunchPattern='sase3'.
[5]:
peaks = tb.get_digitizer_peaks(proposal, runNB, 'FastADC2_9raw', integParams=good_params, bunchPattern='sase3')
peaks
[5]:
<xarray.Dataset> Size: 25MB
Dimensions: (trainId: 7898, sa3_pId: 400)
Coordinates:
* trainId (trainId) uint64 63kB 1501374970 1501374971 ... 1501382869
* sa3_pId (sa3_pId) int32 2kB 772 776 780 784 ... 2356 2360 2364 2368
Data variables:
FastADC2_9peaks (trainId, sa3_pId) float64 25MB -1.04e+04 ... -1.866e+03We see here that peaks has a variable FastADC2_9peaks which has dimensions ['trainId', 'sa3_pId'], which is exactly what we wanted: the raw traces were automatically transformed into vectors of length sa3_pId.
Note that we could also have ommitted the parameter integParams in get_digitizer_peaks() to force the automatic peak finding algorithm:
[6]:
peaks = tb.get_digitizer_peaks(proposal, runNB, 'FastADC2_9raw', bunchPattern='sase3')
If the peak-finding algorithm fails¶
The best strategy is to inspect the raw trace with get_dig_avg_trace() or use check_peak_params() with show_all=True and determine the regions of integration manually. Once the integration parameter dictionnary is created, one can feed it to the integParams argument in get_digitizer_peaks().
Save / load processed peaks¶
If we have found good integration parameters, it is worth saving the integrated peaks as processed data. This can be done by selecting save=True in get_digitizer_peaks(). The location can be chosen by subdir, by default it goes to the usr/processed_runs folder of the proposal.
[7]:
peaks = tb.get_digitizer_peaks(proposal, runNB, 'FastADC2_9raw', integParams=good_params, bunchPattern='sase3',
save=True)
saved data into /gpfs/exfel/exp/SCS/202202/p002956/usr/processed_runs/r0013/r0013-digitizers-data.h5.
To load the processed data:
[8]:
tb.load_processed_peaks(proposal, runNB, 'FastADC2_9peaks')
[8]:
<xarray.DataArray 'FastADC2_9peaks' (trainId: 7898, sa3_pId: 400)> Size: 25MB
array([[-10396.5, -21040.5, -11618. , ..., -2953. , -14307.5, -9120. ],
[-14979.5, -3093.5, -8993. , ..., -5679.5, -4255. , -25531. ],
[-22282. , -2814. , -21852.5, ..., -14205.5, -8499.5, -13072. ],
...,
[ -607.5, 99. , -660. , ..., -1162.5, -1217. , -808. ],
[ -999.5, -681. , -327. , ..., -1800. , -935. , -660. ],
[ -1075. , -1200. , -358. , ..., -753. , -864. , -1866.5]])
Coordinates:
* trainId (trainId) uint64 63kB 1501374970 1501374971 ... 1501382869
* sa3_pId (sa3_pId) int32 2kB 772 776 780 784 788 ... 2356 2360 2364 2368
Attributes:
FastADC2_9peaks_pulseStart: 6763
FastADC2_9peaks_period: 96
FastADC2_9peaks_pulseStop: 6770
FastADC2_9peaks_baseStop: 6762
FastADC2_9peaks_baseStart: 6757
FastADC2_9peaks_npulses: 400Note that the attributes are the peak integration parameters used for peak extraction.
It is also possible to load the entire dataset containing the peaks and average traces of all the processed sources by ommitting the mnemonic argument:
[9]:
tb.load_processed_peaks(proposal, runNB)
[9]:
<xarray.Dataset> Size: 26MB
Dimensions: (trainId: 7898, sa3_pId: 400, sampleId: 100000)
Coordinates:
* trainId (trainId) uint64 63kB 1501374970 1501374971 ... 1501382869
* sa3_pId (sa3_pId) int32 2kB 772 776 780 784 ... 2356 2360 2364 2368
Dimensions without coordinates: sampleId
Data variables:
FastADC2_9peaks (trainId, sa3_pId) float64 25MB -1.04e+04 ... -1.866e+03
FastADC2_9avg (sampleId) float64 800kB -411.8 -412.0 ... -411.3 -411.2
Attributes:
FastADC2_9peaks_pulseStart: 6763
FastADC2_9peaks_period: 96
FastADC2_9peaks_pulseStop: 6770
FastADC2_9peaks_baseStop: 6762
FastADC2_9peaks_baseStart: 6757
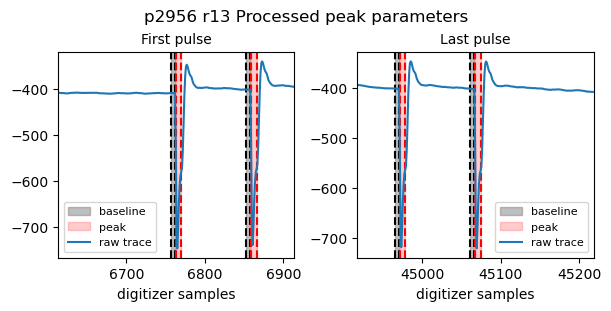
FastADC2_9peaks_npulses: 400It is also possible to check the integration parameters that were used for peak extraction:
[10]:
tb.check_processed_peak_params(proposal, runNB, 'FastADC2_9peaks')
[10]:
{'pulseStart': 6763,
'period': 96,
'pulseStop': 6770,
'baseStop': 6762,
'baseStart': 6757,
'npulses': 400}
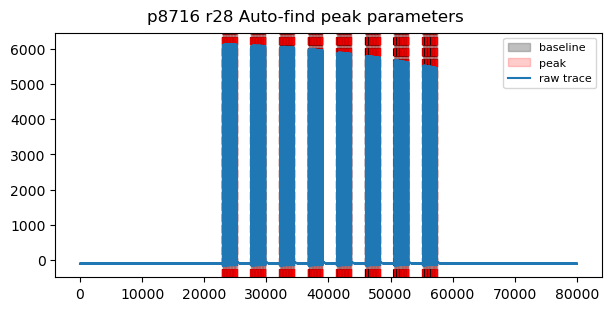
Example of irregular PPL pattern¶
[11]:
# irregular pattern
proposal, runNB, field = 8716, 28, 'I0_ILHraw'
params = tb.check_peak_params(proposal, runNB, field,
bunchPattern='scs_ppl', show_all=True)
Bunch pattern scs_ppl: Not a regular pattern. 192 pulses, pulse_ids=[ 0 4 8 12 16 20 24 28 32 36 40 44 48 52
56 60 64 68 72 76 80 84 88 92 192 196 200 204
208 212 216 220 224 228 232 236 240 244 248 252 256 260
264 268 272 276 280 284 384 388 392 396 400 404 408 412
416 420 424 428 432 436 440 444 448 452 456 460 464 468
472 476 576 580 584 588 592 596 600 604 608 612 616 620
624 628 632 636 640 644 648 652 656 660 664 668 768 772
776 780 784 788 792 796 800 804 808 812 816 820 824 828
832 836 840 844 848 852 856 860 960 964 968 972 976 980
984 988 992 996 1000 1004 1008 1012 1016 1020 1024 1028 1032 1036
1040 1044 1048 1052 1152 1156 1160 1164 1168 1172 1176 1180 1184 1188
1192 1196 1200 1204 1208 1212 1216 1220 1224 1228 1232 1236 1240 1244
1344 1348 1352 1356 1360 1364 1368 1372 1376 1380 1384 1388 1392 1396
1400 1404 1408 1412 1416 1420 1424 1428 1432 1436].
Auto-find peak parameters: Not a regular pattern. 192 pulses, pulse_ids=[ 0 4 8 12 16 20 24 28 32 36 40 44 48 52
56 60 64 68 72 76 80 84 88 92 192 196 200 204
208 212 216 220 224 228 232 236 240 244 248 252 256 260
264 268 272 276 280 284 384 388 392 396 400 404 408 412
416 420 424 428 432 436 440 444 448 452 456 460 464 468
472 476 576 580 584 588 592 596 600 604 608 612 616 620
624 628 632 636 640 644 648 652 656 660 664 668 768 772
776 780 784 788 792 796 800 804 808 812 816 820 824 828
832 836 840 844 848 852 856 860 960 964 968 972 976 980
984 988 992 996 1000 1004 1008 1012 1016 1020 1024 1028 1032 1036
1040 1044 1048 1052 1152 1156 1160 1164 1168 1172 1176 1180 1184 1188
1192 1196 1200 1204 1208 1212 1216 1220 1224 1228 1232 1236 1240 1244
1344 1348 1352 1356 1360 1364 1368 1372 1376 1380 1384 1388 1392 1396
1400 1404 1408 1412 1416 1420 1424 1428 1432 1436].
[ ]: